[ad_1]
Developments in entire genome sequencing have ignited a revolution in digital biology.
Genomics packages internationally are gaining momentum as the price of high-throughput, next-generation sequencing has declined.
Whether or not used for sequencing critical-care patients with rare diseases or in population-scale genetics research, entire genome sequencing is turning into a elementary step in medical workflows and drug discovery.
However genome sequencing is simply step one. Analyzing genome sequencing knowledge requires accelerated compute, knowledge science and AI to learn and perceive the genome. With the end of Moore’s law, the remark that there’s a doubling each two years within the variety of transistors in an built-in circuit, new computing approaches are essential to decrease the price of knowledge evaluation, improve the throughput and accuracy of reads, and in the end unlock the complete potential of the human genome.
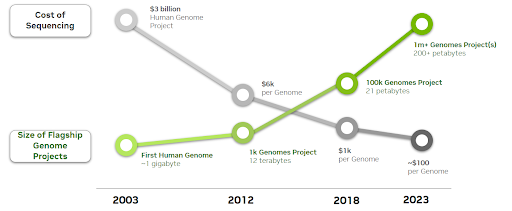
An Explosion in Bioinformatics Information
Sequencing a person’s entire genome generates roughly 100 gigabytes of uncooked knowledge. That greater than doubles after the genome is sequenced utilizing advanced algorithms and purposes resembling deep studying and pure language processing.
As the price of sequencing a human genome continues to lower, volumes of sequencing knowledge are exponentially growing.
An estimated 40 exabytes will probably be required to retailer all human genome knowledge by 2025. As a reference, that’s 8x extra storage than can be required to retailer each phrase spoken in historical past.
Many genome evaluation pipelines are struggling to keep up with the expansive ranges of uncooked knowledge being generated.
Accelerated Genome Sequencing Evaluation Workflows
Sequencing evaluation is sophisticated and computationally intensive, with quite a few steps required to establish genetic variants in a human genome.
Deep studying is turning into vital for base calling proper inside the genomic instrument utilizing RNN- and convolutional neural community (CNN)-based fashions. Neural networks interpret picture and sign knowledge generated by devices and infer the three billion nucleotide pairs of the human genome. That is bettering the accuracy of the reads and guaranteeing that base calling happens nearer to actual time, additional hastening your entire genomics workflow, from pattern to variant name format to closing report.
For secondary genomic evaluation, alignment applied sciences use a reference genome to help with piecing a genome again collectively after the sequencing of DNA fragments.
BWA-MEM, a number one algorithm for alignment, helps researchers quickly map DNA sequence reads to a reference genome. STAR is one other gold-standard alignment algorithm used for RNA-seq knowledge that delivers correct, ultrafast alignment to raised perceive gene expressions.
The dynamic programming algorithm Smith-Waterman can also be extensively used for alignment, a step that’s accelerated 35x on the NVIDIA H100 Tensor Core GPU, which features a dynamic programming accelerator.
Uncovering Genetic Variants
One of the vital important phases of sequencing tasks is variant calling, the place researchers establish variations between a affected person’s pattern and the reference genome. This helps clinicians decide what genetic illness a critically in poor health affected person may need, or helps researchers look throughout a inhabitants to find new drug targets. These variants may be single-nucleotide modifications, small insertions and deletions, or advanced rearrangements.
GPU-optimized and -accelerated callers such because the Broad Institute’s GATK — a genome evaluation toolkit for germline variant calling — improve velocity of research. To assist researchers take away false positives in GATK outcomes, NVIDIA collaborated with the Broad Institute to introduce NVScoreVariants, a deep studying instrument for filtering variants utilizing CNNs.
Deep learning-based variant callers resembling Google’s DeepVariant improve accuracy of calls, with out the necessity for a separate filtering step. DeepVariant makes use of a CNN structure to name variants. It may be retrained to fine-tune for enhanced accuracy with every genomic platform’s outputs.
Secondary evaluation software program within the NVIDIA Clara Parabricks suite of instruments has accelerated these variant callers up to 80x. For instance, germline HaplotypeCaller’s runtime is diminished from 16 hours in a CPU-based surroundings to lower than 5 minutes with GPU-accelerated Clara Parabricks.
Accelerating the Subsequent Wave of Genomics
NVIDIA helps to allow the following wave of genomics by powering each short- and long-read sequencing platforms with accelerated AI base calling and variant calling. Business leaders and startups are working with NVIDIA to push the boundaries of entire genome sequencing.
For instance, biotech firm PacBio lately introduced the Revio system, a brand new long-read sequencing system that includes NVIDIA Tensor Core GPUs. Enabled by a 20x improve in computing energy relative to prior methods, Revio is designed to sequence human genomes with high-accuracy lengthy reads at scale for below $1,000.
Oxford Nanopore Technologies provides the one single expertise that may sequence any-length DNA or RNA fragments in actual time. These options enable the speedy discovery of extra genetic variation. Seattle Youngsters’s Hospital lately used the high-throughput nanopore sequencing instrument PromethION to grasp a genetic dysfunction within the first few hours of a new child’s life.
Ultima Genomics is providing high-throughput entire genome sequencing at simply $100 per pattern, and Singular Genomics’ G4 is probably the most highly effective benchtop system.
Study Extra
At NVIDIA GTC, a free AI convention happening on-line March 20-23, audio system from PacBio, Oxford Nanopore, Genomic England, KAUST, Stanford, Argonne Nationwide Labs and different main establishments will share the latest AI advances in genomic sequencing, evaluation and genomic large language models for understanding gene expression.
The convention contains a keynote from NVIDIA founder and CEO Jensen Huang on Tuesday, March 21, at 8 a.m. PT.
NVIDIA Clara Parabricks is free for college students and researchers. Get started today or try a free hands-on lab to expertise the toolkit in motion.
[ad_2]
Source link



